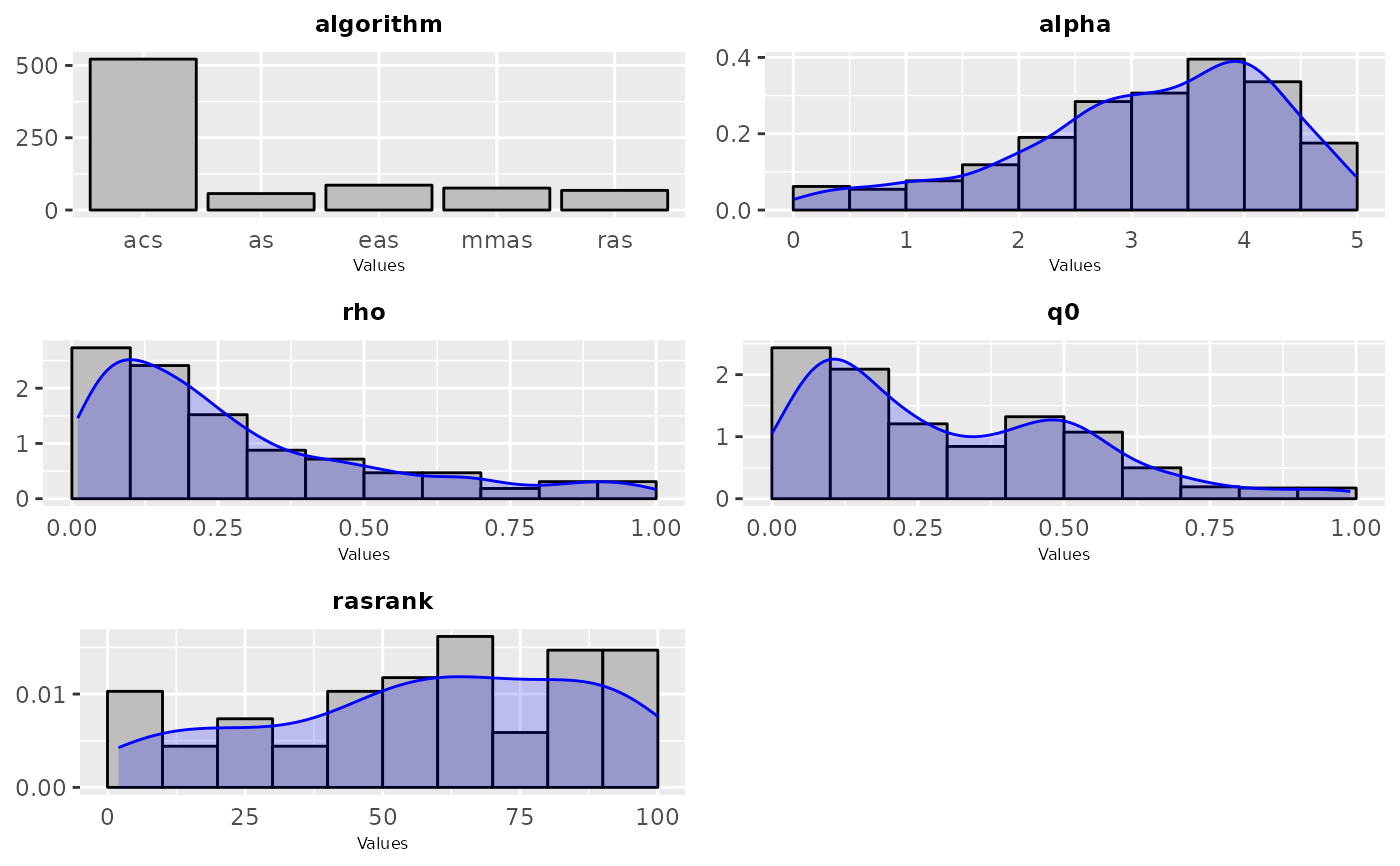
Frequency or density plot that depicts the sampling performed by irace across the iterations of the configuration process. For categorical parameters a frequency plot is created, while for numerical parameters a histogram and density plots are created. The plots are shown in groups of maximum 9, the parameters included in the plot can be specified by setting the param_names argument.
sampling_frequency(
configurations,
parameters,
param_names = NULL,
n = NULL,
filename = NULL
)Arguments
- configurations
(
data.frame())
Configurations iniraceformat. Example:iraceResults$allConfigurations.- parameters
(
list())
Parameters object iniraceformat. If this argument is missing, the first parameter is taken as theiraceResultsdata generated when loading the.Rdatafile created byiraceandconfigurations=iraceResults$allConfigurationsandparameters = iraceResults$scenario$parameters.- param_names
(
character()) Parameters to be included in the plot. Example:c("algorithm","alpha","rho","q0","rasrank").- n
Numeric, for scenarios with large parameter sets, it selects a subset of 9 parameters. For example,
n=1selects the first 9 (1 to 9) parameters, n=2 selects the next 9 (10 to 18) parameters and so on.- filename
(
character(1)) File name to save the plot, for example"~/path/example/filename.png".
Value
Frequency and/or density plot
Note
If there are more than 9 parameters, a pdf file extension is
recommended as it allows to create a multi-page document. Otherwise, you
can use the n argument of the function to generate the plot of a subset
of the parameters.
Examples
# Either use iraceResults
iraceResults <- read_logfile(system.file(package="iraceplot", "exdata",
"guide-example.Rdata", mustWork = TRUE))
sampling_frequency(iraceResults)
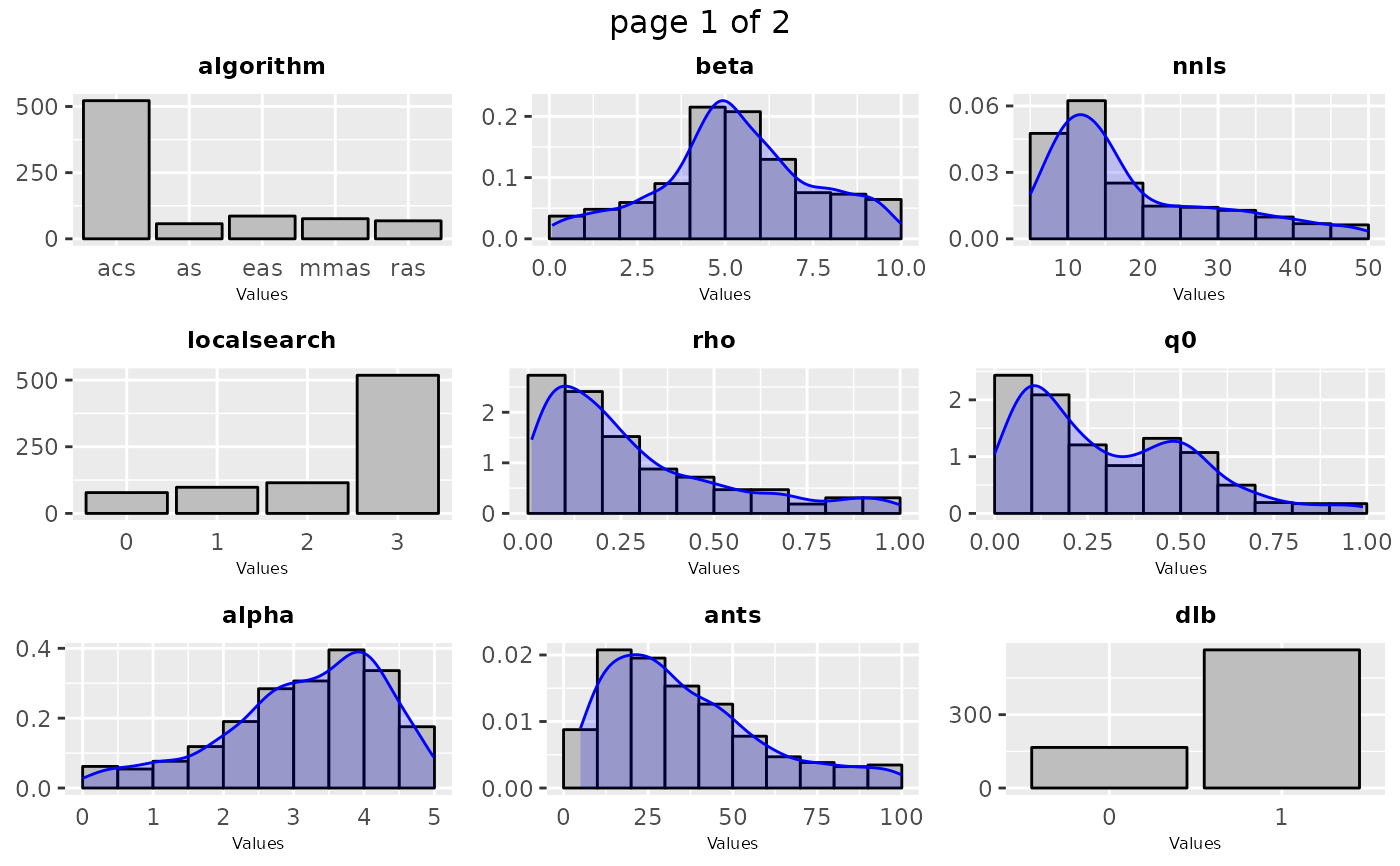
 # \donttest{
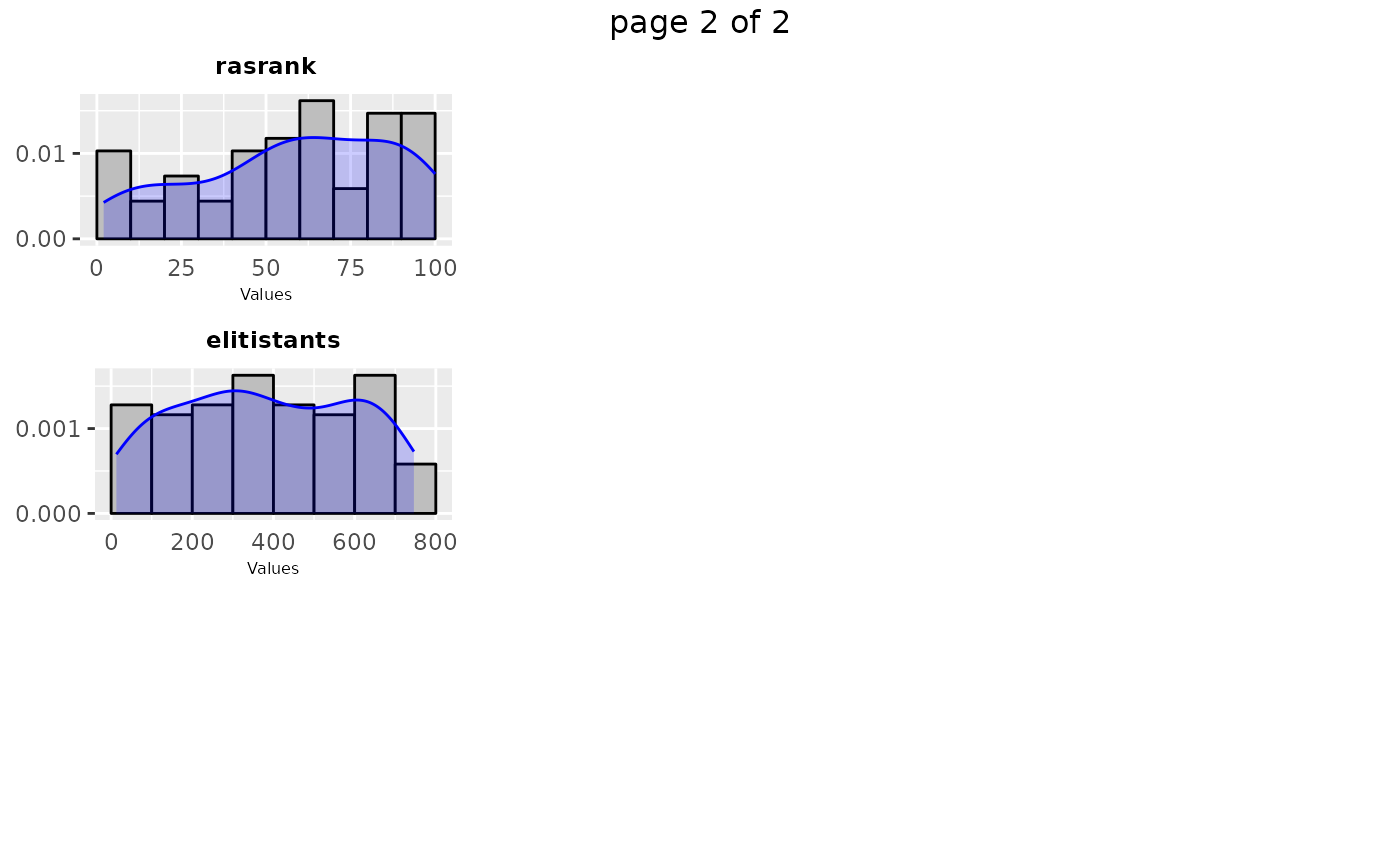
sampling_frequency(iraceResults, n = 2)
# \donttest{
sampling_frequency(iraceResults, n = 2)
 #> TableGrob (2 x 1) "arrange": 2 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (2-2,1-1) arrange gtable[layout]
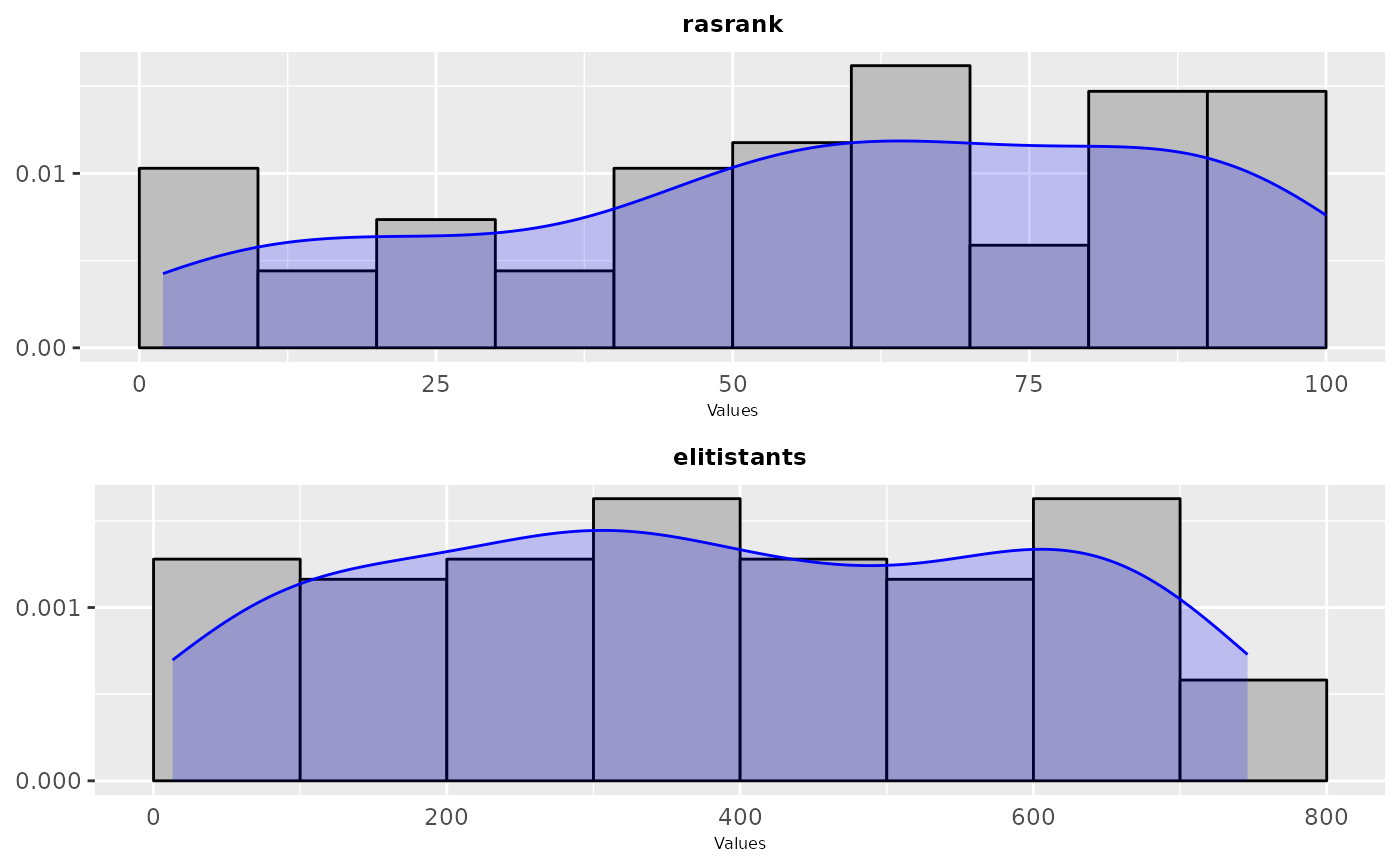
sampling_frequency(iraceResults, param_names = c("alpha"))
#> TableGrob (2 x 1) "arrange": 2 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (2-2,1-1) arrange gtable[layout]
sampling_frequency(iraceResults, param_names = c("alpha"))
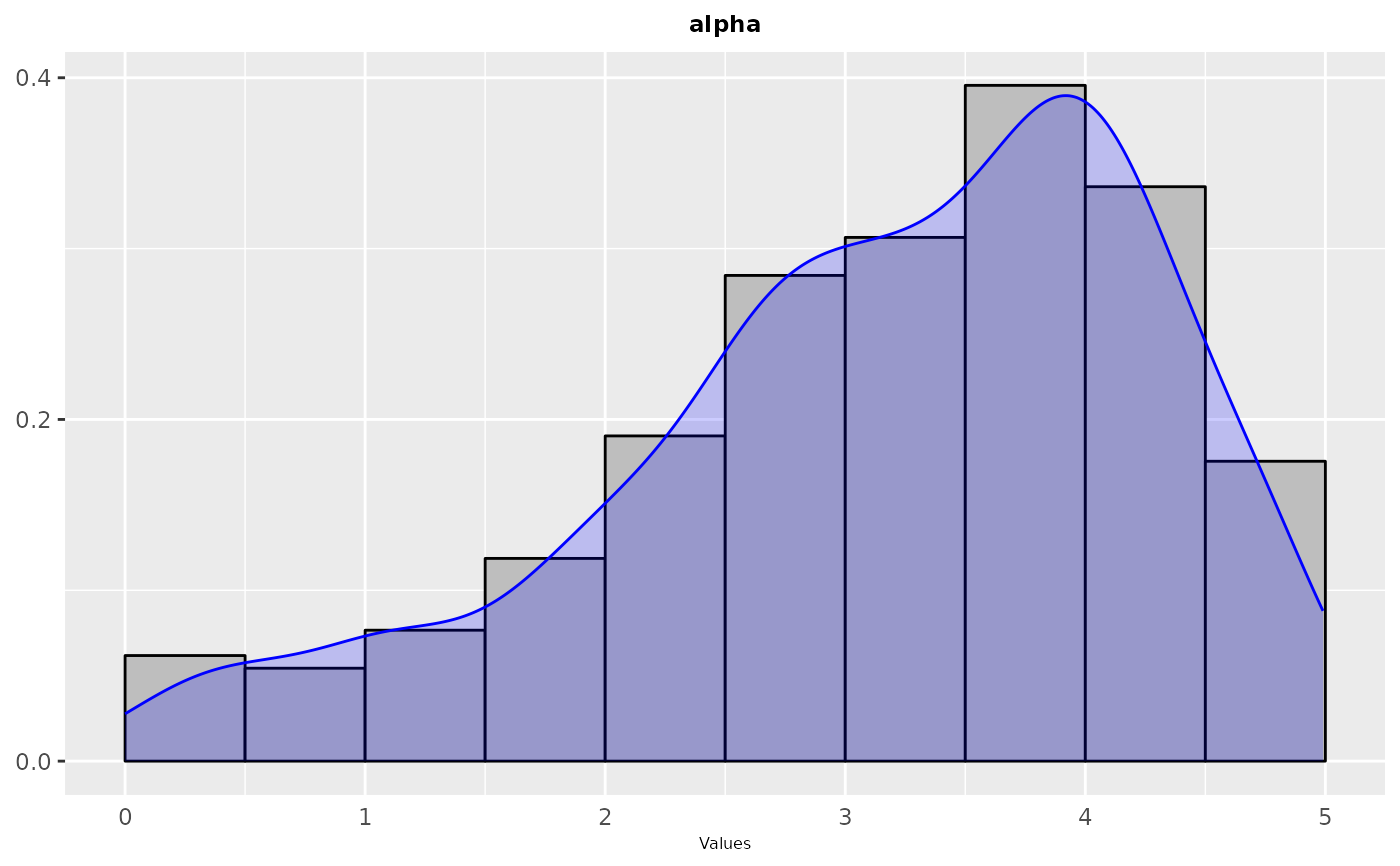 sampling_frequency(iraceResults, param_names = c("algorithm", "alpha", "rho", "q0", "rasrank"))
sampling_frequency(iraceResults, param_names = c("algorithm", "alpha", "rho", "q0", "rasrank"))
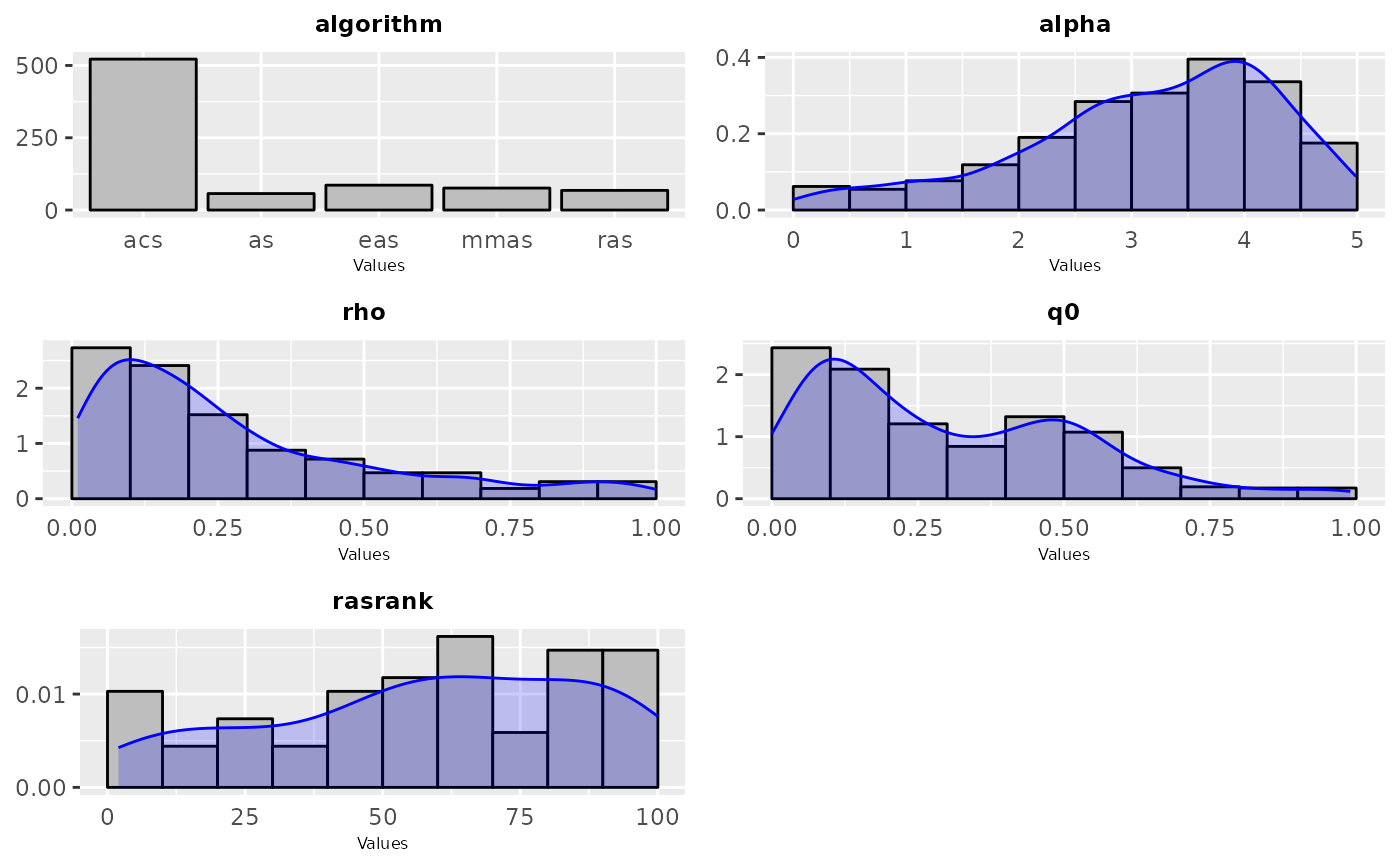 #> TableGrob (3 x 2) "arrange": 5 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (1-1,2-2) arrange gtable[layout]
#> 3 3 (2-2,1-1) arrange gtable[layout]
#> 4 4 (2-2,2-2) arrange gtable[layout]
#> 5 5 (3-3,1-1) arrange gtable[layout]
# }
# Or explicitly specify the configurations and parameters.
parameters <- iraceResults$scenario$parameters
sampling_frequency(iraceResults$allConfigurations, parameters)
#> TableGrob (3 x 2) "arrange": 5 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (1-1,2-2) arrange gtable[layout]
#> 3 3 (2-2,1-1) arrange gtable[layout]
#> 4 4 (2-2,2-2) arrange gtable[layout]
#> 5 5 (3-3,1-1) arrange gtable[layout]
# }
# Or explicitly specify the configurations and parameters.
parameters <- iraceResults$scenario$parameters
sampling_frequency(iraceResults$allConfigurations, parameters)
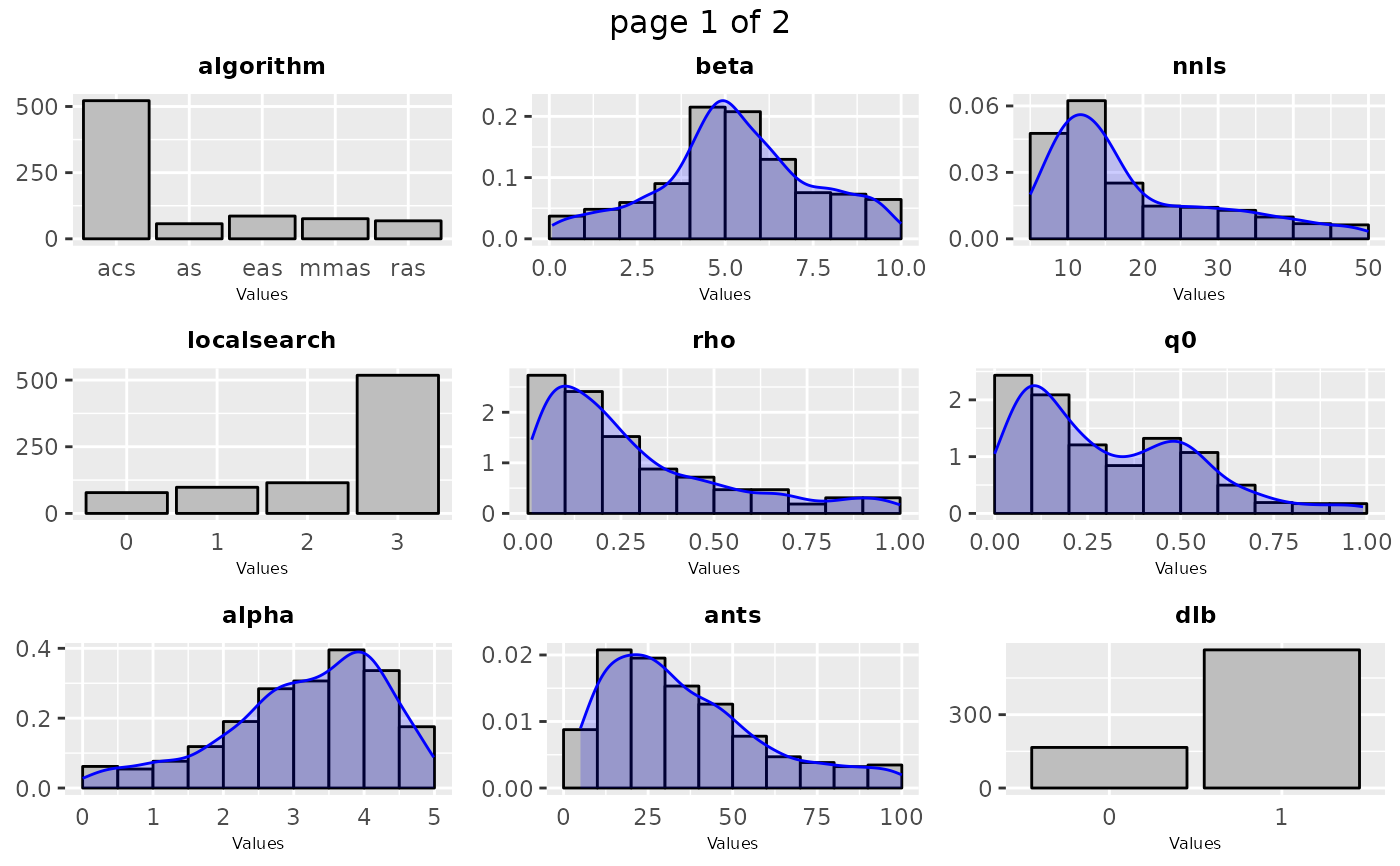
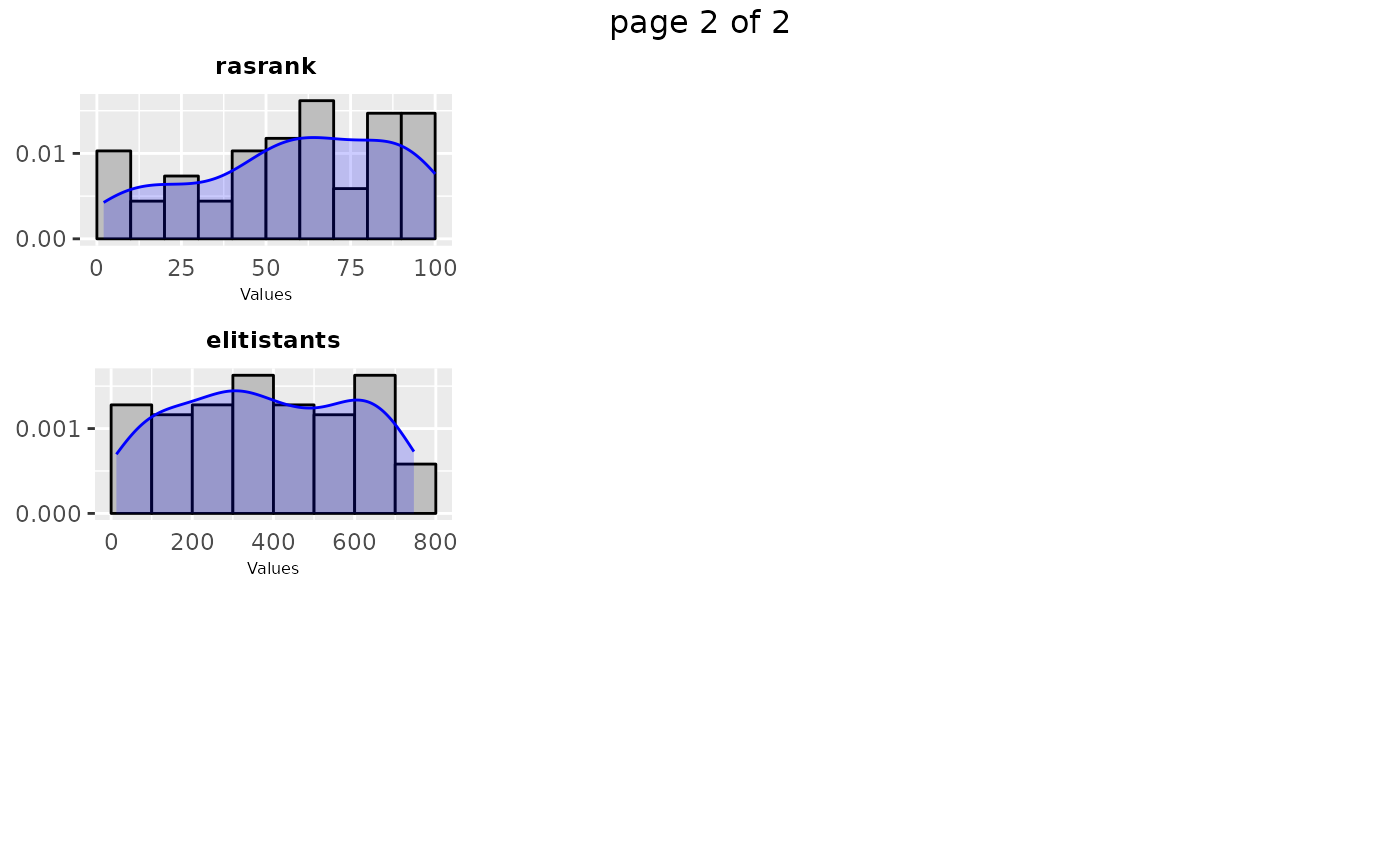 # \donttest{
sampling_frequency(iraceResults$allConfigurations, parameters, n = 2)
# \donttest{
sampling_frequency(iraceResults$allConfigurations, parameters, n = 2)
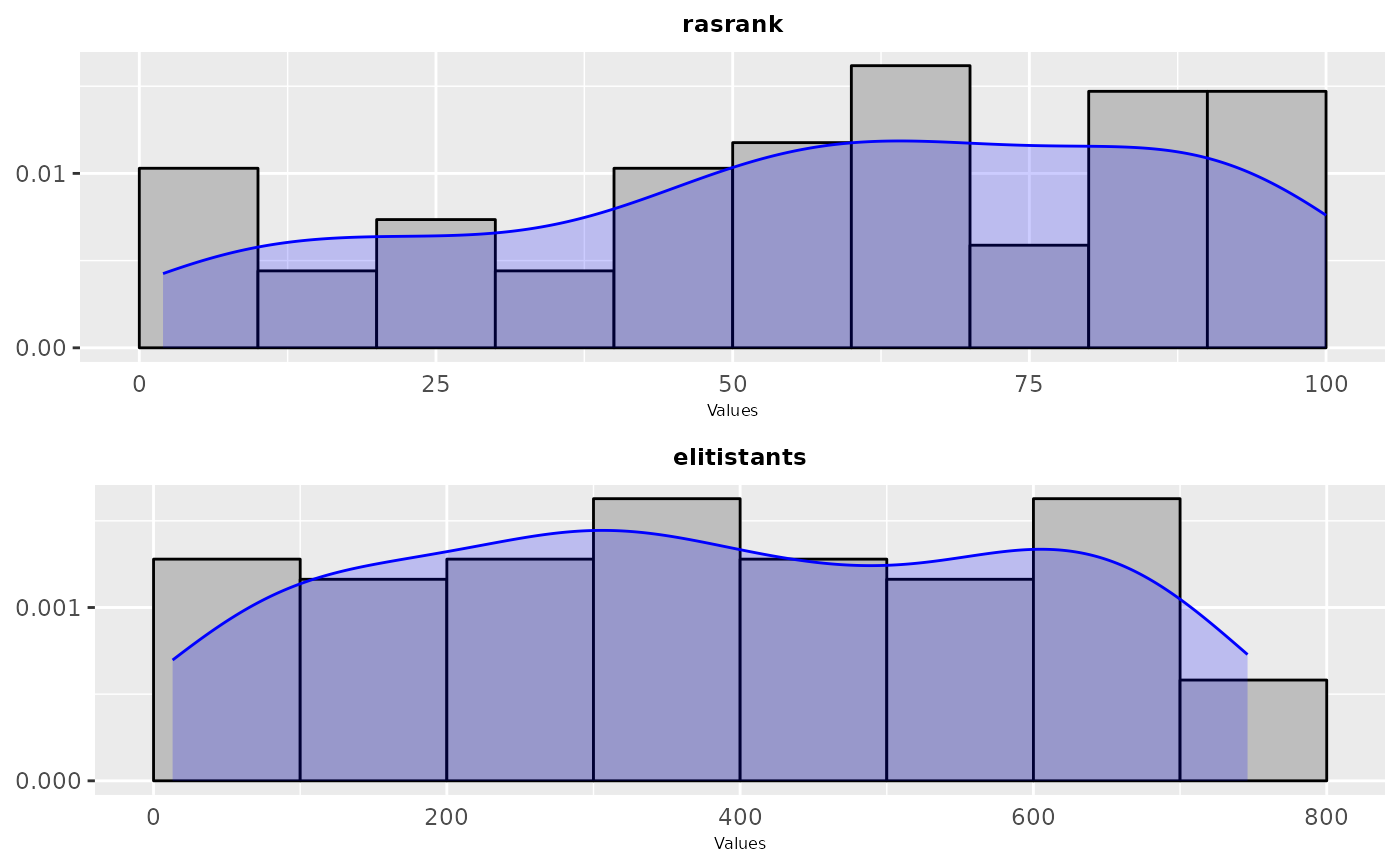 #> TableGrob (2 x 1) "arrange": 2 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (2-2,1-1) arrange gtable[layout]
sampling_frequency(iraceResults$allConfigurations, parameters,
param_names = c("alpha"))
#> TableGrob (2 x 1) "arrange": 2 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (2-2,1-1) arrange gtable[layout]
sampling_frequency(iraceResults$allConfigurations, parameters,
param_names = c("alpha"))
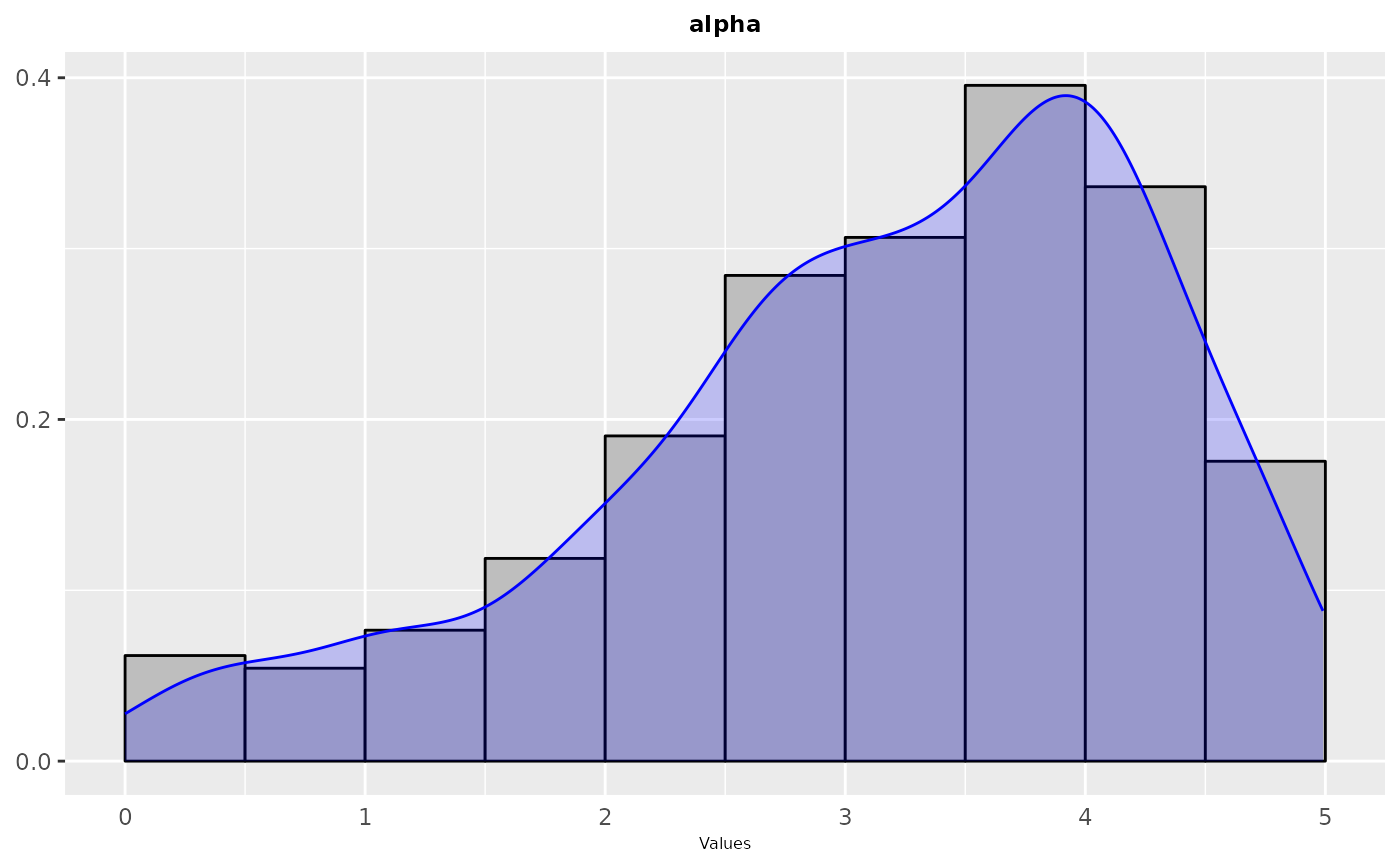 sampling_frequency(iraceResults$allConfigurations, parameters,
param_names = c("algorithm", "alpha", "rho", "q0", "rasrank"))
sampling_frequency(iraceResults$allConfigurations, parameters,
param_names = c("algorithm", "alpha", "rho", "q0", "rasrank"))
 #> TableGrob (3 x 2) "arrange": 5 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (1-1,2-2) arrange gtable[layout]
#> 3 3 (2-2,1-1) arrange gtable[layout]
#> 4 4 (2-2,2-2) arrange gtable[layout]
#> 5 5 (3-3,1-1) arrange gtable[layout]
# }
#> TableGrob (3 x 2) "arrange": 5 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[layout]
#> 2 2 (1-1,2-2) arrange gtable[layout]
#> 3 3 (2-2,1-1) arrange gtable[layout]
#> 4 4 (2-2,2-2) arrange gtable[layout]
#> 5 5 (3-3,1-1) arrange gtable[layout]
# }